Mutation Analyser¶
Eukaryotic genes have coding regions (exons) interrupted by non-coding introns. Exons must be selected and spliced together and introns removed. This process is governed by numerous short exonic sequences called splicing enhancers (ESE) or splicing silencers (ESS). The balance between ESE and ESS determines whether an exons is selected or not. Unsurprisingly, mutations in ESE or ESS sequences can cause human disease due to loss or gain of the exon. Your task is to create an interactive mutation analyser that will help biologists to predict the loss or gain of ESE or ESS sequences.
Task description¶
You can adjust these constraints if you have a good reason. The only rule that you must stick to is: Have something to show on Friday!
- Input
A FASTA file containing a reference sequence and optionally, mutated sequences. An example is
test.fasta- Scoring
Obtain the enhancer/silencer score for each 8-mer along the sequence from
octamers.txt. An 8-mer is an enhancer sequence if both columns (P and I) are above 2.62. It is a silencer if both columns are below -2.62.- Visualisation
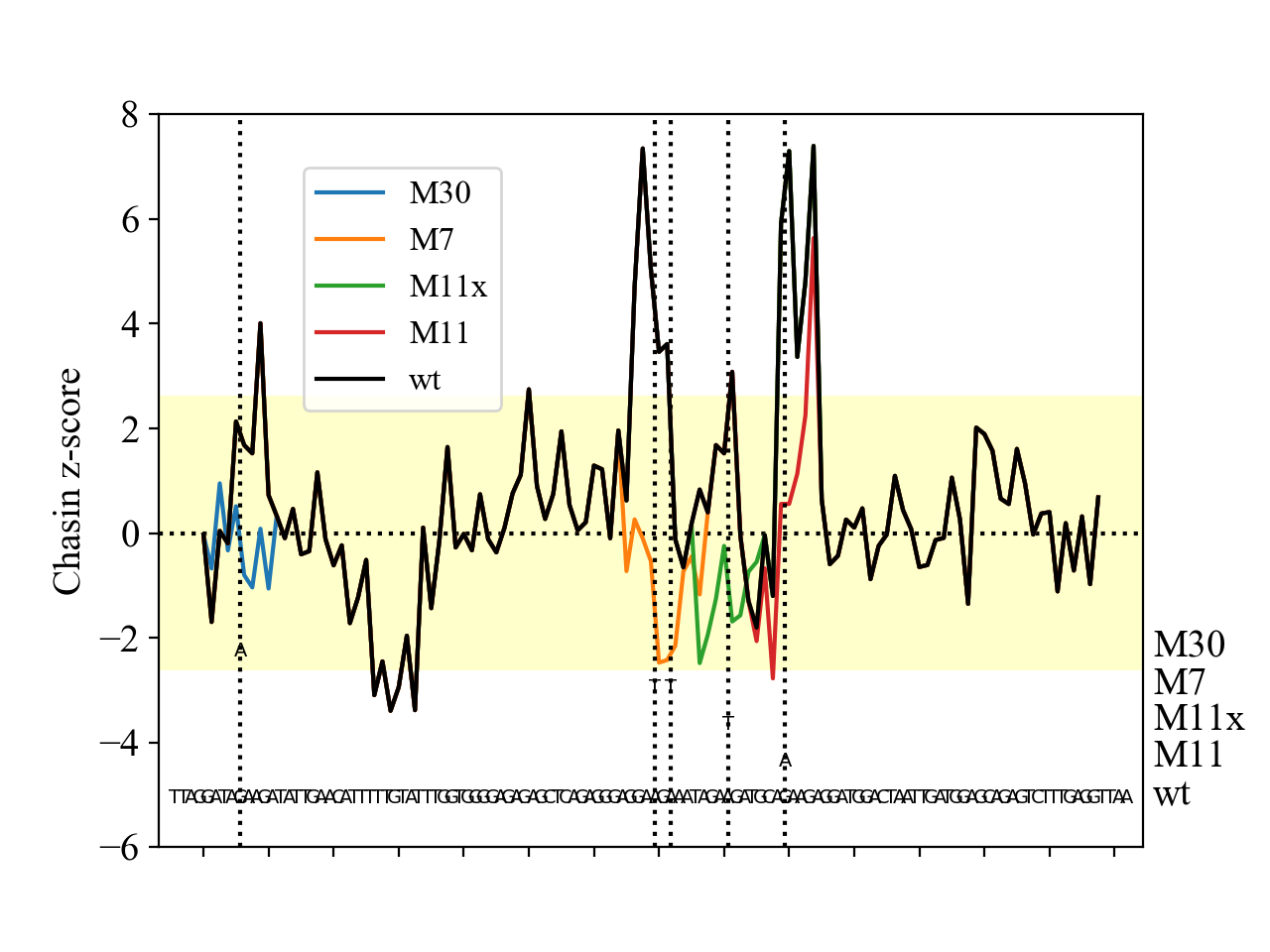
Show a graph of the score plotted over the text of the sequence, as shown below. Find a way of combining the separate P and I scores into a single meaningful plottable value.
- Interactivity
The user should be able to hover the mouse cursor over any of the sequence text and type a different nucleotide letter, with the effect on the graph immediately shown. Use the event handling features in matplotlib.